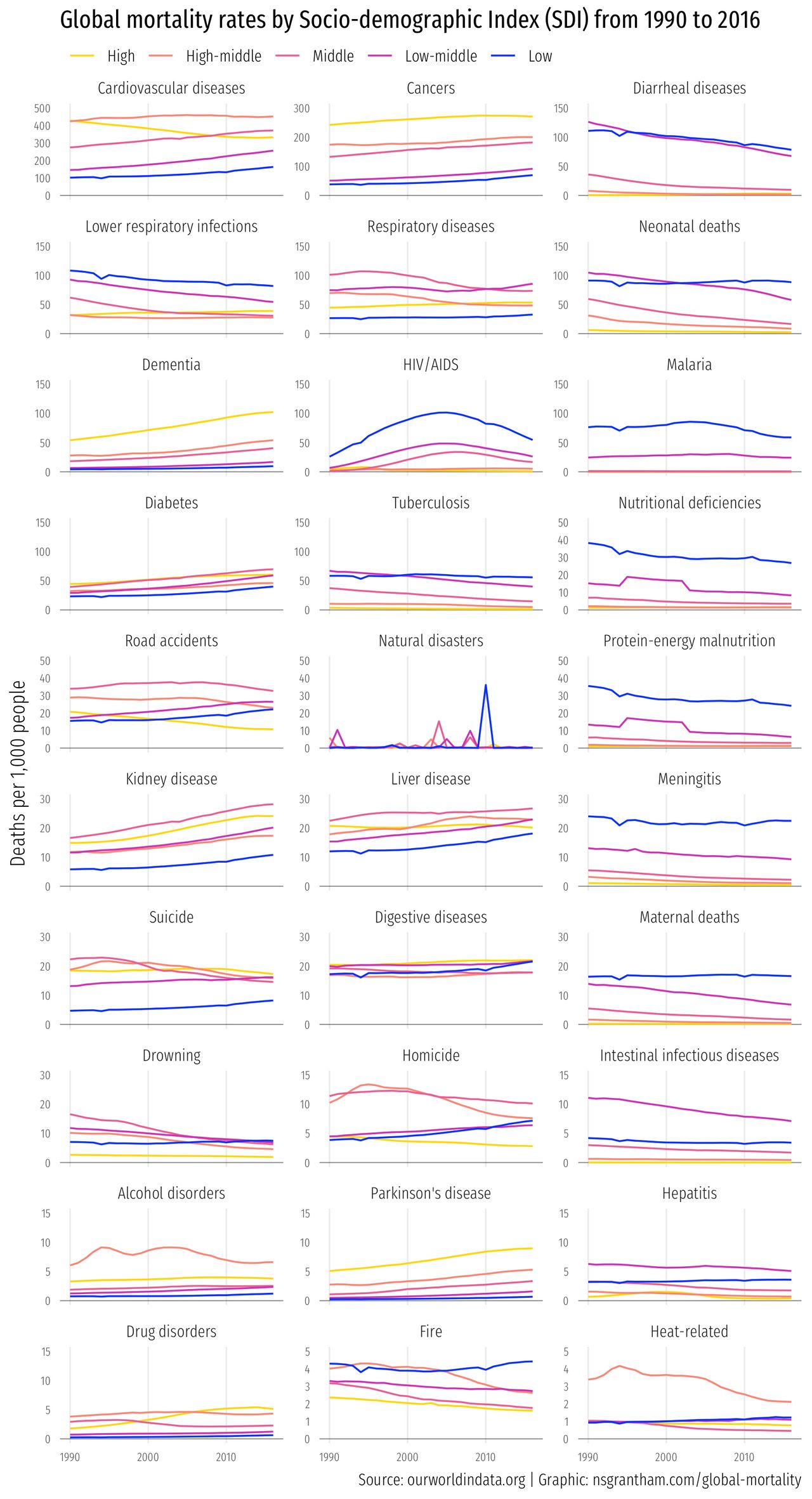
Death comes for us all. And what a splendid variety of forms it can take! Cardiovascular disease, cancer, natural disasters, malaria, infections — the list is almost endless.
This week we’ll look at mortality data collected by the World Health Organization (WHO) to see how cause of death varies by the socio-demographic index of the country in which you live.
library(tidyverse)
library(readxl)
download.file("https://github.com/rfordatascience/tidytuesday/blob/master/data/2018/2018-04-16/global_mortality.xlsx?raw=true", "global_mortality.xlsx")
mortality <- read_xlsx("global_mortality.xlsx") %>%
filter(str_detect(country, "SDI")) %>%
rename(sociodemographic_index = country) %>%
mutate(sociodemographic_index = sub(" SDI", "", sociodemographic_index)) %>%
mutate(sociodemographic_index = fct_relevel(sociodemographic_index, !! c("High", "High-middle", "Middle", "Low-middle", "Low")))
mortality
# A tibble: 135 x 35
sociodemographi… country_code year `Cardiovascular… `Cancers (%)` `Respiratory di…
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 High NA 1990 42.7 24.2 4.45
2 High NA 1991 42.3 24.4 4.47
3 High NA 1992 41.8 24.6 4.50
4 High NA 1993 41.4 24.8 4.56
5 High NA 1994 40.9 25.0 4.59
6 High NA 1995 40.5 25.1 4.65
7 High NA 1996 40.1 25.4 4.70
8 High NA 1997 39.7 25.6 4.76
9 High NA 1998 39.2 25.8 4.81
10 High NA 1999 38.8 25.9 4.89
# … with 125 more rows, and 29 more variables: `Diabetes (%)` <dbl>, `Dementia (%)` <dbl>,
# `Lower respiratory infections (%)` <dbl>, `Neonatal deaths (%)` <dbl>, `Diarrheal diseases
# (%)` <dbl>, `Road accidents (%)` <dbl>, `Liver disease (%)` <dbl>, `Tuberculosis
# (%)` <dbl>, `Kidney disease (%)` <dbl>, `Digestive diseases (%)` <dbl>, `HIV/AIDS
# (%)` <dbl>, `Suicide (%)` <dbl>, `Malaria (%)` <dbl>, `Homicide (%)` <dbl>, `Nutritional
# deficiencies (%)` <dbl>, `Meningitis (%)` <dbl>, `Protein-energy malnutrition (%)` <dbl>,
# `Drowning (%)` <dbl>, `Maternal deaths (%)` <dbl>, `Parkinson disease (%)` <dbl>, `Alcohol
# disorders (%)` <dbl>, `Intestinal infectious diseases (%)` <dbl>, `Drug disorders
# (%)` <dbl>, `Hepatitis (%)` <dbl>, `Fire (%)` <dbl>, `Heat-related (hot and cold exposure)
# (%)` <dbl>, `Natural disasters (%)` <dbl>, `Conflict (%)` <dbl>, `Terrorism (%)` <dbl>
The dataset is in a wide format, so we’ll need to pivot into a longer, tidy format.
mortality <- mortality %>%
select_if(~ all(!is.na(.))) %>%
rename_all(~ sub(" [(]%[)]", "", .)) %>%
gather(cause_of_death, deaths_per_1k, -sociodemographic_index, -year) %>%
mutate(deaths_per_1k = 10 * deaths_per_1k,
cause_of_death = replace(cause_of_death, cause_of_death == "Parkinson disease", "Parkinson's disease"),
cause_of_death = replace(cause_of_death, cause_of_death == "Heat-related (hot and cold exposure)", "Heat-related"))
mortality
# A tibble: 4,050 x 4
sociodemographic_index year cause_of_death deaths_per_1k
<chr> <dbl> <chr> <dbl>
1 High 1990 Cardiovascular diseases 427.
2 High 1991 Cardiovascular diseases 423.
3 High 1992 Cardiovascular diseases 418.
4 High 1993 Cardiovascular diseases 414.
5 High 1994 Cardiovascular diseases 409.
6 High 1995 Cardiovascular diseases 405.
7 High 1996 Cardiovascular diseases 401.
8 High 1997 Cardiovascular diseases 397.
9 High 1998 Cardiovascular diseases 392.
10 High 1999 Cardiovascular diseases 388.
# … with 4,040 more rows
This part looks a little esoteric at first, but the point of it is to calculate an invisible upper limit on a series of facetted plots.
parse_factor_to_numeric <- function(x) as.numeric(levels(x))[x]
yvals <- mortality %>%
group_by(cause_of_death) %>%
summarize(ymax = max(deaths_per_1k),
yupper = cut(max(deaths_per_1k), c(0, 5, 15, 30, 50, 150, 300, 500),
labels = c(5, 15, 30, 50, 150, 300, 500))) %>%
mutate(yupper = parse_factor_to_numeric(yupper)) %>%
arrange(desc(ymax))
mortality <- mortality %>%
left_join(yvals, by = "cause_of_death") %>%
mutate(cause_of_death = factor(cause_of_death, levels = yvals$cause_of_death))
mortality
# A tibble: 4,050 x 6
sociodemographic_index year cause_of_death deaths_per_1k ymax yupper
<chr> <dbl> <fct> <dbl> <dbl> <dbl>
1 High 1990 Cardiovascular diseases 427. 459. 500
2 High 1991 Cardiovascular diseases 423. 459. 500
3 High 1992 Cardiovascular diseases 418. 459. 500
4 High 1993 Cardiovascular diseases 414. 459. 500
5 High 1994 Cardiovascular diseases 409. 459. 500
6 High 1995 Cardiovascular diseases 405. 459. 500
7 High 1996 Cardiovascular diseases 401. 459. 500
8 High 1997 Cardiovascular diseases 397. 459. 500
9 High 1998 Cardiovascular diseases 392. 459. 500
10 High 1999 Cardiovascular diseases 388. 459. 500
# … with 4,040 more rows
Lookin’ good.
Now we’ll make line graphs with year on the x-axis, deaths per 1,000 people on the y-axis, and lines colored by the socio-demoographic index (SDI) from ‘Low’ to ‘High’.
ggplot(mortality, aes(year, deaths_per_1k, color = sociodemographic_index)) +
geom_hline(yintercept = 0, size = 0.3, alpha = 0.5) +
geom_hline(aes(yintercept = yupper), size = 0.3, alpha = 0) +
geom_line() +
facet_wrap(~ cause_of_death, scales = "free_y", ncol = 3) +
scale_color_manual(values = c("#ffd700", "#fa8775", "#ea5f94", "#cd34b5", "#0000ff")) +
labs(title = "Global mortality rates by Socio-demographic Index (SDI) from 1990 to 2016",
subtitle = "",
caption = "Source: ourworldindata.org | Graphic: nsgrantham.com/global-mortality",
x = NULL, y = "Deaths per 1,000 people") +
theme_minimal(base_family = "Fira Sans Extra Condensed Light", base_size = 12) +
theme(plot.title = element_text(family = "Fira Sans Extra Condensed"),
legend.position = c(0.33, 1.0358),
legend.direction = "horizontal",
legend.title = element_blank(),
axis.text = element_text(size = 7),
panel.grid.major.y = element_blank(),
panel.grid.minor.y = element_blank(),
panel.grid.major.x = element_line(size = 0.4),
panel.grid.minor.x = element_blank())
ggsave("global-mortality.png", width = 6.5, height = 12)
Boy, what a heart-warming graphic.